Overview of my research:
Peptide Conformational Ensemble Generation via Generative Model (in progress)
So far, sophisticated protein structure prediction methods have emerged. However, they are primarily limited to folded proteins and do not offer comprehensive insights into equilibrium properties. In this research, I aim to develop a generative model capable of producing an equilibrium ensemble of proteins. This project is supported by JST ACT-X.
Constant-froce Steered Molecular Dynamics Simulation
We proposed the constant-force steered molecular dynamics simulation to estimate unbiased dissociation rate.
The input structures and topologies are provided 👉 github.
Our review paper for non-equilibrium molecular dynamics simulation 👉 Iida, S. & Tomoshi, K. Free energy and kinetic rate calculation via non-equilibrium molecular simulation: application to biomolecules.
Cryptic Binding Site of Proteins
We have found unique side-chain fluctuations of cryptic binding sites via molecular dynamics simulations.
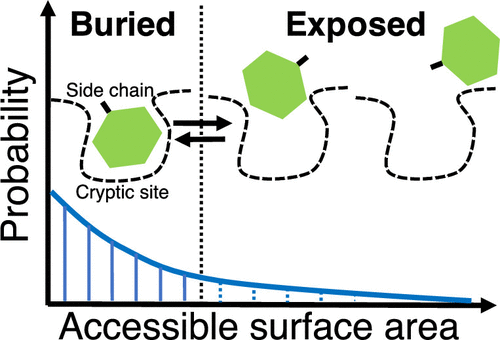
Source code to analyse the fluctuations 👉 github
Intrinsically Disordererd Protein
We have explored the structural ensemble of an intrinsically disordered region, p53 C-terminal domain, via a generalised ensemble molecular dynamics simulation. We have identified various binding modes on a target protein.
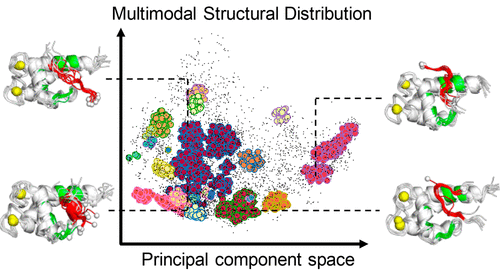
Our papers 👇
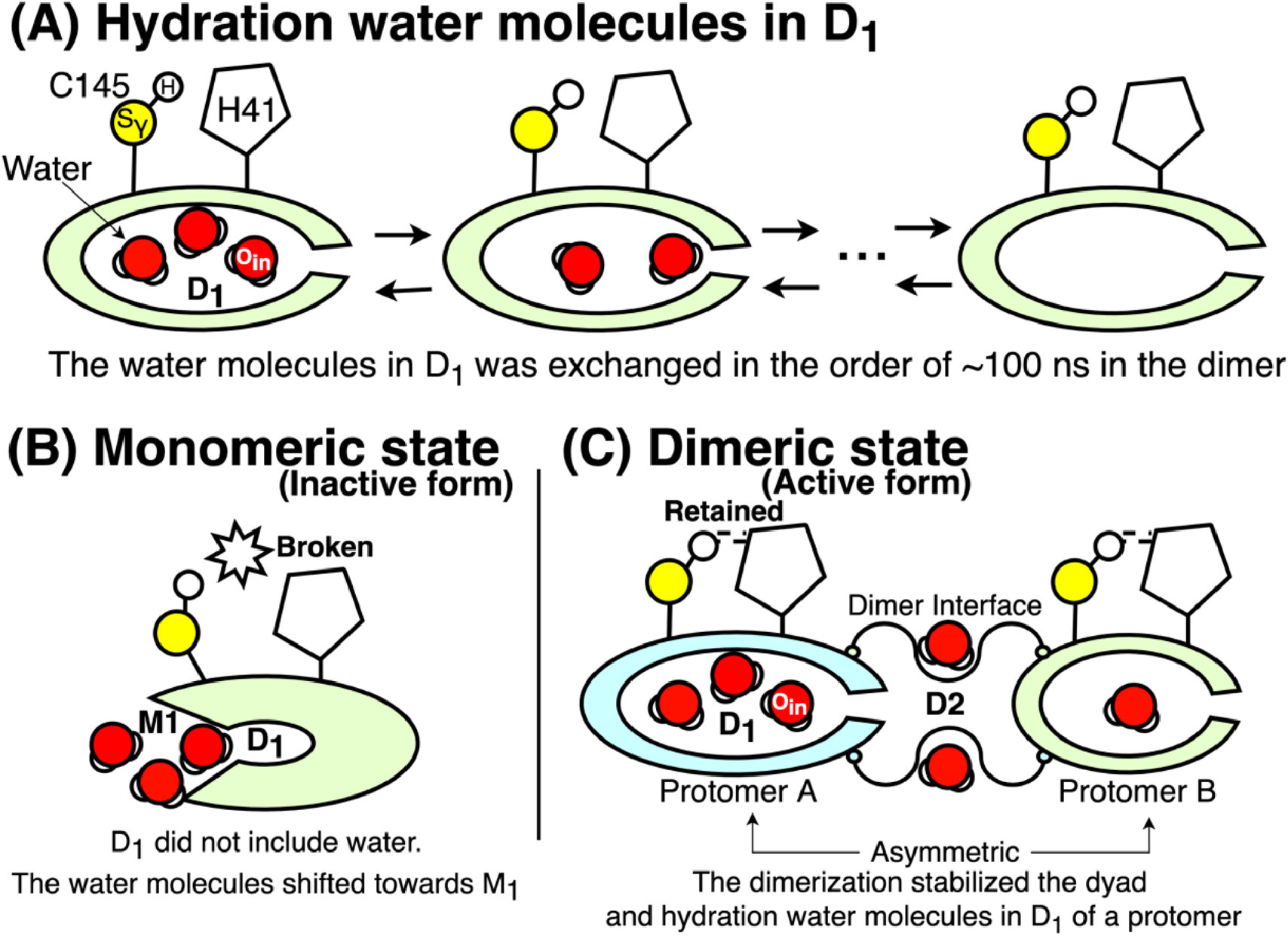
SARS-CoV-2 main protease
We have investigated monomeric and dimeric states of the SARS-CoV-2 main protease, identifying water molecules that can be crucial to keep the dyad configuration.

 Google Scholar
Google Scholar Twitter
Twitter  DockerHub
DockerHub